Digital PCR – a technology set to transform clinical testing?
on LinkedIn:
Pregnancy screening, cancer treatment, organ transplant – digital PCR testing has the power to enhance clinical decision-making. But what is needed to take it mainstream?
Polymerase Chain Reaction (PCR) testing has reached unlikely levels of fame due to the COVID-19 pandemic. However, its latest evolution, digital PCR, could be a real game-changer for commercial diagnostics.
The power of digital PCR
When people talk about PCR testing, they often refer to quantitative PCR. This technology is fantastic at delivering binary answers, for example, whether a disease is present or not. It can also determine to some extent how much of a disease is present in a sample (though the process is inaccurate). Quantitative PCR’s quantification can be improved by calibrators, but this is complex, expensive, and time-consuming for a lab to perform.
Digital PCR advances this technology to deliver precise quantification and improves detection of low-frequency DNA targets.
The potential to transform clinical decision-making
The key benefit of digital PCR can be summed up in two words: better data. It has the power to transform clinical decision-making, for example, in the following areas:
Pregnancy screening
Highly accurate testing for chromosomal trisomies, such as Down’s syndrome, by detecting traces of foetal DNA in maternal blood. Next-generation sequencing (NGS) is an existing alternative but has extremely complex protocols, including DNA purification, DNA library preparation, sequencing, data alignment, and analysis.
Cancer treatment
Pinpointing disease progression by detecting tumor DNA in liquid biopsies.
Organ transplants
Detecting DNA sequences leaking from a donor organ (a sign that the host immune system is rejecting it).
Virus detection
Increasing accuracy in treatment of HIV, hepatitis C, herpes, cytomegalovirus, and other infections.
Disease diagnostics
Quantification of bacterial species in stools due to digital PCR’s lower sensitivity to inhibitors.
Digital PCR could also deliver accurate quantification of levels of other infectious diseases such as respiratory viruses and sexually transmitted infections. This precision isn’t currently available but could be useful for clinicians to differentiate between different stages of infection.
How does digital PCR differ from quantitative PCR?
Digital PCR is a development of ‘standard’ PCR, using the same concept of exponential amplification of template DNA with DNA primers and a polymerase enzyme. It has two crucial differences: compartmentalization and end-point data collection.
Compartmentalization
Instead of performing a reaction on a whole sample, digital PCR splits the sample across a large number of separate compartments. ‘Compartment’ could mean a microfluidics chip, or a droplet suspended in an emulsion.
Each reaction is capable of detecting a single molecule of DNA. A larger number of amplification cycles are generally run, typically 60 versus 40 for standard PCR. Just one DNA molecule in a compartment is enough to initiate a PCR amplification reaction.
End-point data collection
Unlike quantitative PCR which reads after every amplification cycle, digital PCR just needs to read once when all the amplification cycles are complete. This is an important saving, as otherwise, all the thousands of individual compartments would need to be read every cycle, which would be a significant challenge.
Digital PCR overview
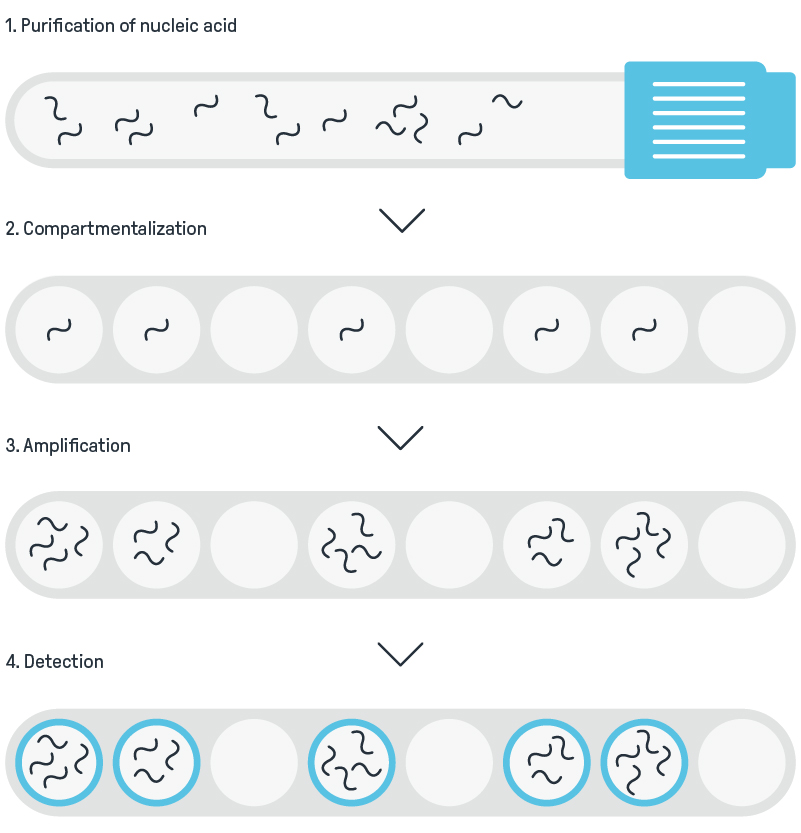
What’s stopping the mass adoption of digital PCR?
Though digital PCR has been around for 20 years and is mentioned in thousands of patents, only a handful of commercial products use the technology. The primary challenge innovators need to crack for it to go mainstream is optimal compartmentalization.
Cracking compartmentalization
Compartmentalization affects key performance parameters, such as the assay’s dynamic range, linearity, accuracy and ease of use; its cost; whether it’s run as a batch or on-demand; and how many samples can be run at once. The number of compartments in the assay must be high, relative to the concentration of input DNA molecules in the sample. But if the assay uses too few compartments, the accuracy of quantification will be too low, and the assay must be repeated using a diluted sample.
Why does compartment design matter?
Digital PCR’s randomly apportioned target molecules across a large number of compartments mean there will be some compartments with no targets, some with one, and a few with two or more. As it’s not known how many target molecules are in each positive compartment, the Poisson distribution is used to determine the most likely proportions of compartments with one, two, three, or more DNA targets. Using the Poisson distribution allows accurate quantification, but relies on two important factors:
- The input template being randomly spread throughout all the reaction chambers
- All compartments being the same size
These are critical parameters, and there are two main ways to achieve them. The first is passing the sample over a microfluidic flow cell containing microwells commonly filled using capillary action. The second is encapsulating the nucleic acid in a huge number of water droplets in an emulsion of oil, with each droplet containing a separate reaction.
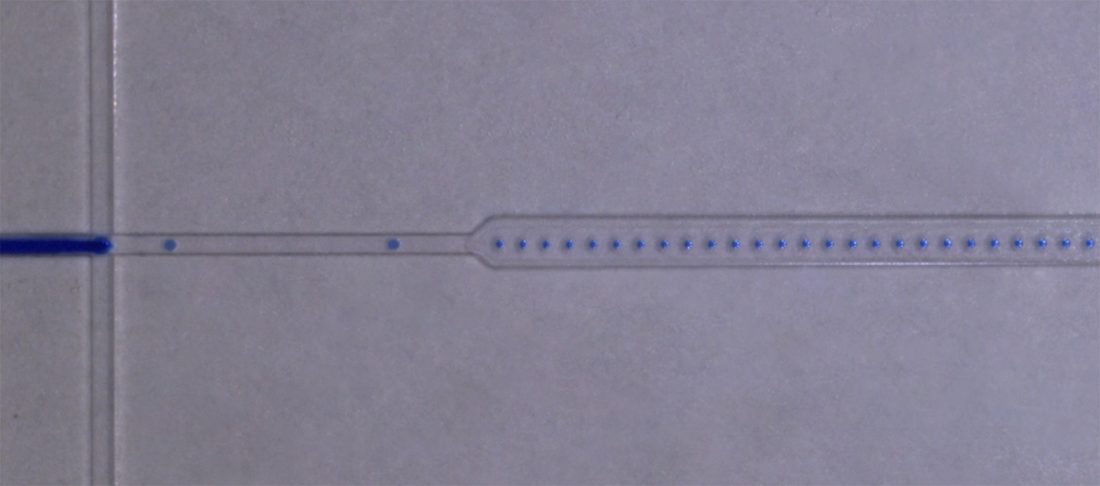
Microfluidic droplet generator developed at CDP
The ideal compartmentalization system would retain the ease of use of current quantitative PCR systems and have a similar lab-bench footprint and costs. However, current approaches (typically involving microfluidics or droplets), require multiple complex disposables and sophisticated optics. If a new approach was developed with lower costs, clinicians and test centers may well convert to digital PCR for all their applications. The company that manages to crack this challenge has the potential to dominate the PCR market and provide huge advances to clinical decision making.
To talk to us about our current innovation in the field of digital PCR, get in touch.

Richard Owen
Principal Bio-Scientist